Rossmann-toolbox server
Only sequence method is available at this moment, stay tuned for structure predictorSubmit:
Please provide a full-length protein sequence in which a cofactor-binding core will be detected with either DeepRossmann or HHpred. Alternatively, turn off the core detection procedure and provide only the core region sequence.
Please provide a structure in the PDB format and the name of the chain in which a cofactor-binding core will be detected with either DeepRossmann or HHpred.
More complex jobs can be run with the standalone version available at our GitHub.
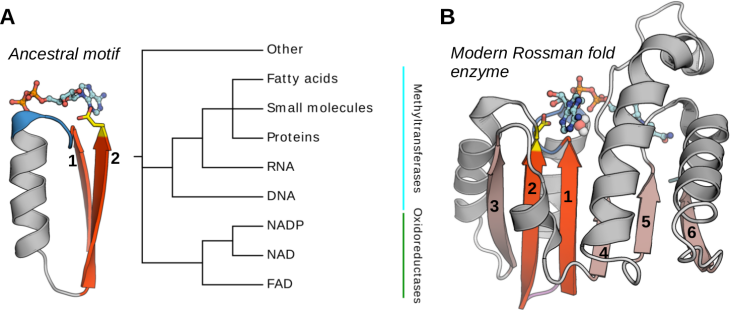
The Rossman fold is one of the most prominent folds in Protein Data Bank and by far the most functionally diverse one, with >300 different functions, including methyltransferases and various oxidoreductases involved in most of the essential cellular pathways. It is also assumed to be one of the oldest folds, which was already well represented in the last universal common ancestor (LUCA).
From the structural perspective, the Rossmann fold belongs to the general class of β/α proteins and comprises four connecting α-helices and six consecutive β-strands (arranged in the 3-2-1-4-5-6 order) forming a parallel pleated sheet. Rossmann-fold enzyme families are characterized by their use of cofactors and in particular of nucleoside-containing cofactors such as S-Adenosylmethionine (SAM), nicotinamide adenine dinucleotide (NAD), nicotinamide adenine dinucleotide phosphate (NADP), flavin adenine dinucleotide (FAD), and others. These cofactors share not only the biochemical compound (adenosine) but also bind to the same specific region of the Rossmann fold, even in distantly related proteins.
The cofactor-binding site shared by all members of the Rossmann fold ("core") corresponds to a small structural fragment comprising β1–α1–β2 and the connecting loops.
In our study, we showed that despite its short length the βαβ motif unambiguously defines the specificity towards the cofactor. Following this observation, we trained two complementary deep learning models for the prediction of the cofactor specificity based on the features of the βαβ motif. First, DeepLigand, utilizes contextualized sequence embeddings, whereas the second, DeepLigand 3D, relies on structures represented as graphs. A benchmark on two test sets, one containing βαβ motifs bearing no resemblance to those of the training set, and the other comprising 38 cases of the experimentally confirmed redesign of the cofactor specificity from NAD to NADP and vice versa, revealed nearly-perfect performance (95% accuracy) of the two methods.
For more information visit our help page!